The list of molecules of the year is out now at C&E News (but you have to have an account to view the list, unlike previous years).♣ These three caught my eye:
- Electron in a cube: Synthesis and characterization of perfluorocubane as an electron acceptor,[1]. I have already written about this system and will not discuss it further, except to note this one topped the poll!
- Vernier template synthesis of molecular knots[2]
- Megalo-Cavitands: Synthesis of Acridane[4]arenes and Formation of Large, Deep Cavitands for Selective C70 Uptake[3]
The last two are examples of large three-dimensional molecules with unusual properties. The second is an example of a trefoil-of-trefoils, called a triskelion knot and I was very keen to get hold of its coordinates so that I could inspect the knotting. I thought I might summarise here the hierarchical procedures one might try for acquiring such data.
- The most modern method of acquiring data associated with an article is to inspect the citation list at the end of the article. The trend encouraged by the FAIR data principles suggests that if such data has an associated DOI (as indeed the article itself does), then this DOI should be cited in the citations just like articles themselves. This concept is also known as treating data as a first class citizen of the scholarly processes. In this case no data was associated with the 81 citations listed at 10.1126/science.abm9247
- The prevalent method since ~1996 has been to next download any ESI. That is linked here. I cannot help but note that the PDF format is not one optimised for data, but its better than nothing. This PDF has 114 pages, and one eventually finds the following on p 103: structures and corresponding energies uploaded to the Github database (https://github.com/kjhstenlid/AshbridgeVernier2022/). Github is known as a software repository, but its use as a data repository is unusual. Thus no DOI is assigned this data (which would explain why its not listed in the article citations). Here one learns from the readme that it contains Molecular knot structures in cif-file format for the Verner and Sheild knots.
- To get this data one has to pretend it is code, and download the ZIP code archive. The CIF file found there however gives a fatal error when trying to load into a CIF viewer such as Mercury: Reading cell from Cif failed, could not retrieve ‘_cell_length+a’. The CIF is clearly not generated from a crystallographic analysis program but a modelling program and is clearly invalid as a CIF.
- One now has to fall back seeing if the CIF file can be rescued using a text editor. This is non-trivial but about 10 minutes of editing finally produces a file that can be viewed.
- Here is the 3D structure (click on the image to view).

Now for the Megalo-Cavitands (or not). Just as above, one ends up in a 49-page PDF file looking for coordinates. There one gets pictures of PM6-computed models starting on p 28, but alas apparently no associated coordinates.†
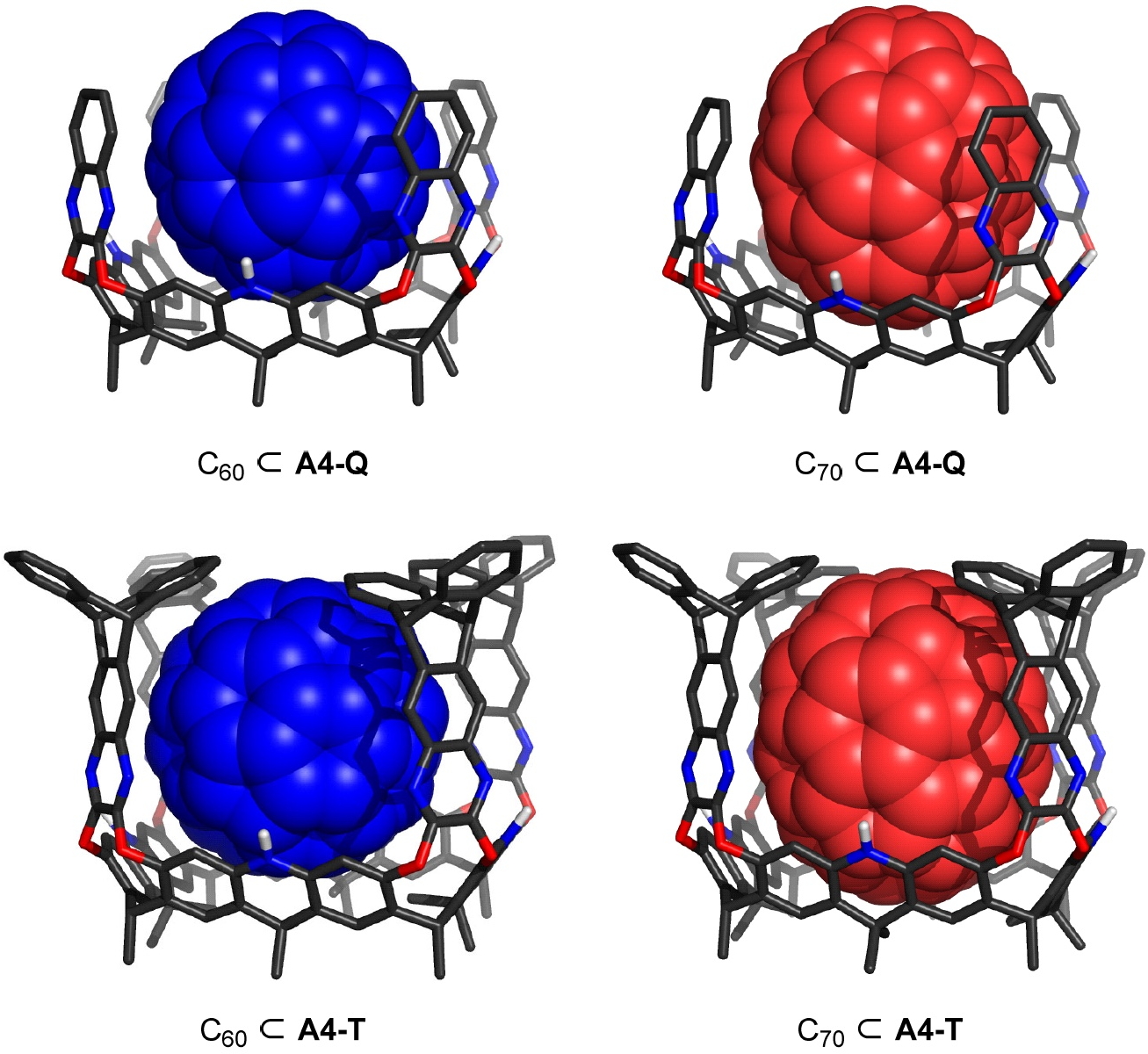
So no 3D models to show here then (sorry, clicking on the image above will not produce them†).
My concluding remark should be that when an interesting molecule is selected for inclusion in eg the molecules of the year – 2022, one of the criteria for its inclusion is that the availability of full and FAIR data describing its properties should be one of the essential criteria for selection.
‡I note the method used to generate these coordinates (PM6) is perhaps not ideal; it contains no dispersion attraction terms, which are probably important if modelling host-guest complexation. The PM7 method which does is far better for this sort of thing! This highlights the importance of providing data, in this case 3D coordinates. It would be interesting to recompute the dimensions of these molecules using a method that does allow for dispersion attractions to be included. For just such an example, see here.
† I have contacted the authors of [3] and it turns out a reference to a Data repository submission was omitted from the article. The data is at DOI: 10.5281/zenodo.6953961 and I will report separately on my analysis of the effect of replacing PM6 with PM7.
♣See this open letter about changes at C&EN.
This post has DOI: 10.14469/hpc/12028
References
- M. Sugiyama, M. Akiyama, Y. Yonezawa, K. Komaguchi, M. Higashi, K. Nozaki, and T. Okazoe, "Electron in a cube: Synthesis and characterization of perfluorocubane as an electron acceptor", Science, vol. 377, pp. 756-759, 2022. https://doi.org/10.1126/science.abq0516
- Z. Ashbridge, E. Kreidt, L. Pirvu, F. Schaufelberger, J.H. Stenlid, F. Abild-Pedersen, and D.A. Leigh, "Vernier template synthesis of molecular knots", Science, vol. 375, pp. 1035-1041, 2022. https://doi.org/10.1126/science.abm9247
- J. Pfeuffer‐Rooschüz, S. Heim, A. Prescimone, and K. Tiefenbacher, "Megalo‐Cavitands: Synthesis of Acridane[4]arenes and Formation of Large, Deep Cavitands for Selective C70 Uptake", Angewandte Chemie International Edition, vol. 61, 2022. https://doi.org/10.1002/anie.202209885