I want to describe a recent attempt by a group of collaborators to share the research data associated with their just published article.[1]
I am here introducing things in a hierarchical form (i.e. not necessarily the serial order in which actions were taken).
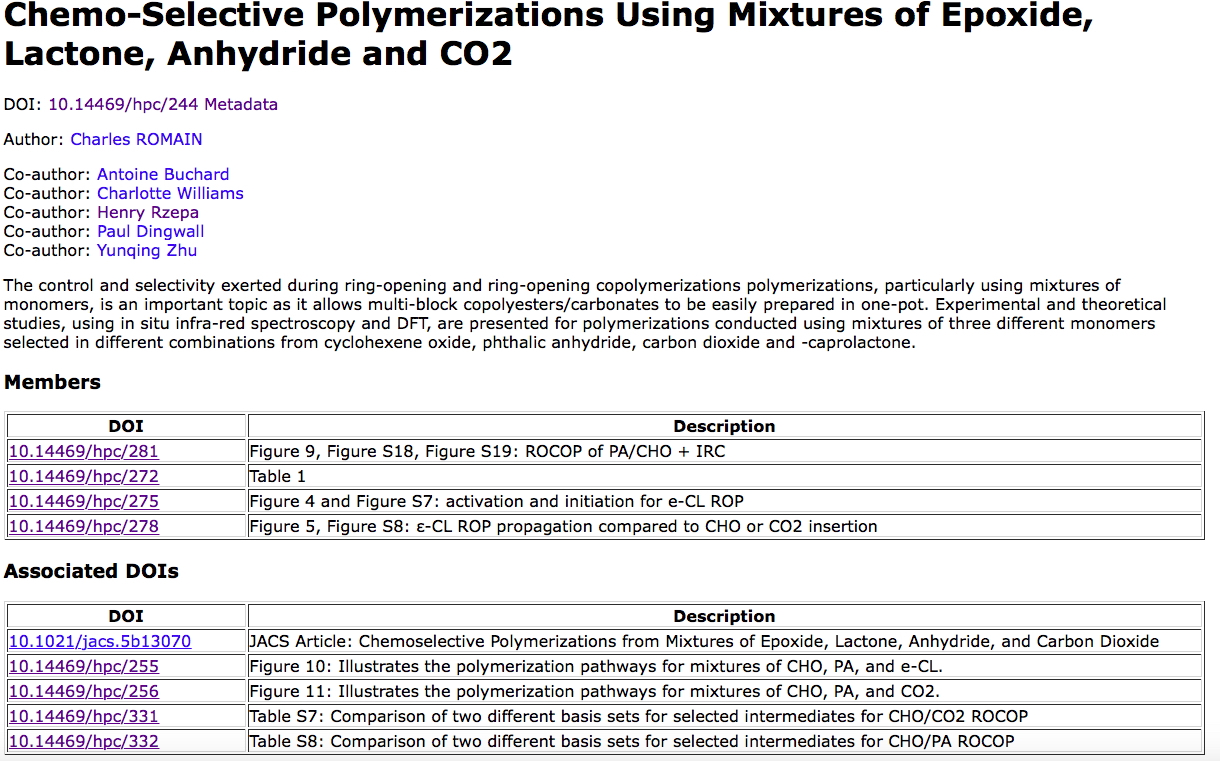
- The data repository selected for the data sharing is described by (m3data) doi: 10.17616/R3K64N[2]
- A collaborative project collection was established on this repository (doi: 10.14469/hpc/244[3]). This data collection has some of the following attributes:
- Its metadata is sent here: https://search.datacite.org/ui?&q=10.14469/hpc/244 where it can be queried for other details.
- The project collaborators are all identified by their ORCID, used to obtain further individual information about the researchers. This information is also propagated to the metadata sent to DataCite.
- In the section labelled associated DOIs there is a link to the recently published peer-reviewed article, which itself cites the data via doi: 10.14469/hpc/244 and which thus establishes a bidirectional link between the article and its data.
-
Also in the associated DOIs section are other DOIs (to two figures and two tables) held in a separate location. One example: doi: 10.14469/hpc/332[4]) which illustrates the original type of data sharing we started about 10 years ago. This form has been variously called a "WEO" or Web-enhanced object (by the ACS) or interactivity boxes (RSC, etc). In such WEOs, we wrap the data into an interactive visual appearance using Jmol or JSmol software. The data itself is directly available to the reader using the Jmol export functions (right mouse click in the visual window).
- In this specific example the WEO has been assigned its DOI using the repository noted above.[2]
- We have in the past also used Figshare[5]) for this purpose, see e.g. 10.6084/m9.figshare.1181739‡
- The WEO itself can itself reference a more complete set of data used to create the visual appearance, for example data that allows the wavefunction of the molecule to be computed, doi: 10.6084/m9.figshare.2581987.v1[6] In this instance this is held on the Figshare[5] repository.
-
The collection has another section labelled Members. These are individual datasets associated with the collection and held on the SAME repository as the collection itself. In this case, there are five such members, two of which are listed below:
- 10.14469/hpc/281[7] contains a variety of other data such as outputs from an IRC (intrinsic reaction coordinate), energy profile diagrams and ZIP archives of other calculations.
-
10.14469/hpc/272[8] itself contains five members, one of which is e.g.
- 10.14469/hpc/267[9] which contains a ZIP archive with NMR data (see here for how this might be packaged in the future) and a file for a GPC (chromatography) instrument.
- This last item also contains a new section labelled Metadata, which includes e.g. the InChI key and InChI string for the molecule whose properties are reported.
If this mode of presenting data seems a little more complex than a single monolithic PDF file, its because its designed for:
- collaboration between scientists, potentially at different locations and institutions.
- attribution of provenance/credit for the individual items (via ORCID).
- separate date stamping by the various contributors.
- providing bi-directional links between data and publications.
- holding what we call FAIR (findable, accessible, interoperable and reusable) data, rather than just data encapsulated in a PDF file.
- Collecting, storing and sending metadata for aggregation in a formal way, i.e. to DataCite using a formal schema to render the metadata properly searchable.
Thus 10.14469/hpc/244 represents our most complex attempt yet at such collaborative FAIR data sharing with multiple contributors. The tools for packaging many of the datasets are still quite limited (see again here) and the design is still being optimised (call it α). When the repository[2] has been more extensively tested, we intend to make it available as open source for others to experiment with. And of course, when this happens the source code too will have its own DOI!
‡A refactoring of the Figshare site in December 2015 meant that the DOI no longer points directly to the WEO, and you have to follow a manually inserted link on that page to see it.
References
- C. Romain, Y. Zhu, P. Dingwall, S. Paul, H.S. Rzepa, A. Buchard, and C.K. Williams, "Chemoselective Polymerizations from Mixtures of Epoxide, Lactone, Anhydride, and Carbon Dioxide", Journal of the American Chemical Society, vol. 138, pp. 4120-4131, 2016. https://doi.org/10.1021/jacs.5b13070
- Re3data.Org., "Imperial College Research Computing Service Data Repository", 2016. https://doi.org/10.17616/r3k64n
- C. ROMAIN, "Chemo-Selective Polymerizations Using Mixtures of Epoxide, Lactone, Anhydride and CO2", 2016. https://doi.org/10.14469/hpc/244
- H. Rzepa, "Table S8: Comparison of two different basis sets for selected intermediates for CHO/PA ROCOP.", 2016. https://doi.org/10.14469/hpc/332
- Re3data.Org., "figshare", 2012. https://doi.org/10.17616/r3pk5r
- P. Dingwall, "Gaussian Job Archive for C6H10O", 2016. https://doi.org/10.6084/m9.figshare.2581987.v1
- C. ROMAIN, "Figure 9, Figure S18, Figure S19: ROCOP of PA/CHO + IRC", 2016. https://doi.org/10.14469/hpc/281
- C. ROMAIN, "Table 1 : Polymerizations Using Lactone, Epoxide, and CO2", 2016. https://doi.org/10.14469/hpc/272
- C. ROMAIN, "Table 1, entry 1 : Polymerizations Using Lactone, Epoxide, and CO2", 2016. https://doi.org/10.14469/hpc/267
Tags: 10.17616, Academic publishing, DataCite, energy profile diagrams, Figshare, Identifiers, Open science, ORCiD, PDF, Scholarly communication, Technical communication, Technology/Internet, Web-enhanced object