We have been experimenting with full-colour 3D printing of molecular objects. I thought I might here share some of our observations. Firstly, I list the software used:
- Crystal structures as sources of ball&stick models (e.g. the CCDC database).
- Gaussian style cube files for sources of wavefunctions.
- There are plenty of other sources of course, in fact any file type which is supported by ..
- Jmol 13.3 (the app, not the applet) to render the above.
- And www.shapeways.com who provide the full colour 3D printing service.
My first attempt was to 3D print a molecular orbital; in fact the one shown in this post. To be printable, a 3D object must be fully connected, in other words not contain any disconnected components. To ensure this is true for a molecular orbital, one has to select a very low isosurface threshold; 0.005 au in this case. The orbital is computed as a Gaussian cube, and converted to a .wrl file as follows
- Start the Jmol.app.
- In the console, enter e.g.
isosurface sign color yellow green cutoff 0.005 "144.cub"
(where 144.cub is the Gaussian cube file). Then write model.wrl
to write out a VRML (.wrl) file.- Create an account at Shapeways, go to “make your own” and upload the .wrl file. After about five minutes, it should appear under “my models“, looking something like below:

- Select “full color sandstone” as the material and see what the quoted price is. For the above it was €3.25. It is actually quite tricky to anticipate what physical size this represents, but upon printing and receiving it by post, it turned out to be about 2 cm in size! I should point out that it takes a day or so for Shapeways to confirm that the model has been sent to manufacturing (it is perfectly possible that it may be rejected at this stage because it cannot be manufactured for whatever reason).
- The thing now is to get the model to scale. To do this, a scale-factor has to be applied to the VRML file, which is easier said than done. Fortunately (thanks Bob Hanson!) this can be applied at the Jmol stage, and after step 2 above in the Jmol console, apply the command:
set exportScale 8.0
to scale up by 8-fold, before writing out the .wrl file. You know it has worked when the size is given as cm: 13.228 w x 13.288 d x 2.786 h and the price goes up to €69. I will assume (I have not printed at this scale yet) that it is less than 13 cm across, but something big enough to hand around a group of people. I should mention that we tried a variety of ways of scaling the model using programs such as Blender, but they all proved unreliable. You can of course also set the colours in the command above. - It is possible to set the scale larger than 8, but say at 10-fold the materials available to you reduce significantly. Full-colour sandstone is no longer available, and you can only print in a single colour (the materials by the way include stainless steel, for €5700.71).
- The resulting model is made from gypsum with a polymer binder. I have not had the courage yet to test how robust it is by breaking one of the models; I suspect it might be fairly fragile. Time will tell.
- I also tried a ball&stick. The molecule selected has been described in another post and is saved by converting the .cif file to .pdb (using in my case the program Mercury). This helps normalise the bond types, producing:
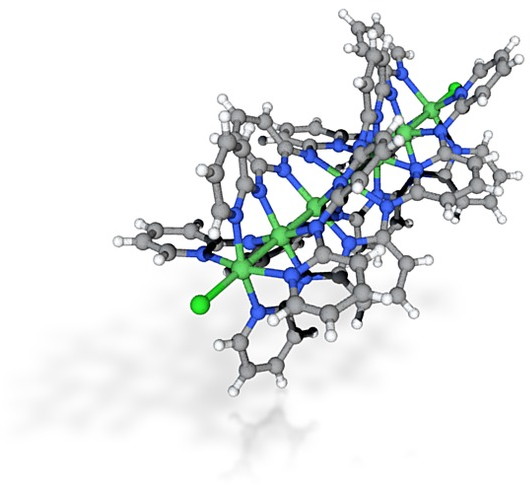
- In this case, converting the crystal coordinates via Jmol to .wrl required a scaling factor of 10.0 in this instance to produce a model costed as €24.53. It is very much a matter of adjusting the
exportscaleagainst the resulting printing price to get the desired result (by trial and error, but perhaps there is a more systematic way of doing this?). One of my colleagues (Paul) has had this sort of thing printed to about 25cm in size for about this price, and the model seems reasonably robust to physical handling. It may also benefit by fine-tuning of the bond radius; thicker would clearly be stronger.‡
If anyone reading this post has their own experience of 3D colour printing of chemical models, do please post comments here. And I dare say that in a few years time, students will simply press the “3D print” button on the tablet they are using to view lecture notes to get a copy. Mind you, I am somewhat ambivalent about such a process, having spent the last twenty years trying to discourage students from using the “2D print” button on their computer. Will I eventually come to adopt the same attitude to 3D print (if you click on the MO image above, you will get a virtual 3D model instead of a physical one).
‡ The width of a bond can be set using the Jmol command: set bondRadiusMilliAngstroms 300
Acknowledgments
This post has been cross-posted in PDF format at Authorea.
Tags: 3D colour printing, 3D printing, Bob Hanson, chemical models, EUR
Can we see some photos of the 3D models?
Here are three different color combinations of the MO above, next to a computer mouse to give the scale. As I noted, I might try the 16 cm version next.
Dean Tantillo alerted me to this article: doi 10.1021/ed3000364, which uses 3D printing in chemistry as an aid for blind or visually impaired people.
The 3D printed models I noted in my post included surfaces (wavefunctions) and ball&stick (including spacefill) representations of covalent (connected) molecules. Many other multidimensional properties also suggest themselves (we wrote about this a few years back; O. Casher, C. Leach, C. S. Page and H. S. Rzepa, Chem. Brit., 1998, 34, 26).
Here I speculate whether 3D-printable models should be routinely included in journal articles, and if so, how to do so? For example, in the post I included two links: http://shpws.me/oTLF and http://shpws.me/oTLB which will take you to the Shapeways page, where (for payment) you can request that the 3D printed model be sent to you. There may be a time quite soon when most organisations will have a 3D-print service, and you will not need to wait a week or so for the model to be shipped to you.
You can instead simply download the .wrl file at that site (for free, it’s a public file) and view it locally (I have verified this works with the two files above).
Or, one might be a devil’s advocate and ask whether what we use already (Jmol, JSmol for example) is already good enough and that 3D print, like 2D print, may already be on its way out of this increasingly virtual world (if you can load it on to e.g. an iPad, why print it?).
So, does the future of chemistry (publishing) reside in virtual or real 3D objects? If they latter, should they be printable?
Would it be possible to print X ray diffraction data of proteins? This would be very useful for examining active sites and binding of small molecules.
I can see two problems with 3D printing X-ray data
1. The object cannot have components suspended in empty space.
2. To be useful for a protein, the printed object would have to be large (> 50 cm?). This is likely to be very expensive!
For most purposes one wouldn’t need to print the whole protein, just the region of interest (active cleft or binding surface).
If there is a component floating in space it isn’t likely to be attached to the protein.
A common symptom of submitting 3D models for printing is that the mesh is invalid for some reason. The following software: http://www.netfabb.com seems to perform a useful service in validating and repairing models. It also comes in a pro-version for power users. You will have to save the model in Jmol as
.objrather than.wrlNick, I think yes, you could get a protein –or part of it– printed from X-ray data. That’s what I’d be interested on, too.
The major issues I see are:
1- Deciding the style of rendering of the protein: molecular surface? balls and stick? The results in printed form may critically depend on this, in terms of validity of the model mesh, robustness of the product, etc., but also the feeling or degree of information you will get from the physical model.
2- You need some mastery of Jmol to prepare the model conveniently –and avoid loose bits, too narrow stretches, …
3- Quite a deal of trial and error — time if not money!!
I am gradually tuning the settings. For example, to print the HOMO molecular orbital of butadiene, the following seem better than the commands given above:
>zap
>load bd_mol15.cub
>select all
>cpk 200
>set bondradiusmilliangstroms 500
>isosurface sign color yellow green cutoff 0.02 bd_mol15.cub
>set exportscale 12.0
>write bd_mol15.wrl
I think this sort of optimisation will have to be performed for most models.
Lee Cronin has reported 3D-printed reactionware with printed-in catalysts.
Here’s a fun idea: Catalyst printed into a model of the catalyst.
Just pluck the model of the catalyst you want to use out from the display cabinet and dunk into the reaction. When your done, simply pluck the model out of the reaction mixture, give it a rinse and a little shake, and place back in the display cabinet.
By the way, Shapeways accept the more modern X3D file format, which can also be written by Jmol.
> write X3D “fileName”
Re: X3D, yes they accept a variety of formats. But I can see no obvious benefit of using X3D vs WRL; they seem entirely equivalent (and so they should, X3D is simply an XML serialisation of WRL).
Do you have a doi for Cronin’s work?
The idea of a library (cabinet) is good. We actually have such a cabinet in the entrance to our chemistry dept. It contains 3D models of molecules, but embedded in lead crystal glass (and in monochrome). No doubt some of the models I have printed will find their way in!
Integrated 3D-printed reactionware for chemical synthesis and analysis: 10.1038/nchem.1313
Six π-MOs from benzene: http://shpws.me/oYY1 http://shpws.me/oYY6 http://shpws.me/oYYa (occupied) http://shpws.me/oYYh http://shpws.me/oYYk http://shpws.me/oYYp (unoccupied) and four from cis-butadiene: http://shpws.me/oYZa http://shpws.me/oYZd (HOMO), http://shpws.me/oYZn (LUMO), http://shpws.me/oYZp were generated following the procedure outlined above (the costs relate purely to manufacturing, with no percentage given to the designer!). They will be used in my undergraduate lectures (time will tell whether they are robust enough for an enthusiastic student audience).
Henry,
Do you think it is feasible to use MRI, CAT and X-ray to create accurate 3-D models of hip joints (or indeed any other joint) and using carbonated compounds or structurally comparable biomaterials recreate via 3D-printing? Would ceramic derivatives be compatible with the molecular 3D printing process and would the chemical bonds created be sufficiently strong to manage body weight and the full range of movement (wear and tear etc). Many thanks.
I can only answer the previous question by saying the learning curve is actually both steep and extensive. We have looked at a variety of different machines used to 3D print in a variety of materials. Modern printers nowadays can manufacture in several materials, either consecutively (depositing one on the other) or at the same time. They can even blend a continuously varying mixture of polymers.
I cannot answer your questions, but I do strongly suspect that this area (like other technologies such as eg mobile phones) is going to advance very rapidly indeed over the next decade.
Can you lay out color setting commands in the Console. I think we can try making a model in Chem3D from Chemdraw and save the file into .PDB file which can be opened and saved as .wrl in JMol. What i can’t do is to render colors for each atom/atom types. Help would be appreciated.
Thanks
Mahender
Atom colours can be set using the standard Jmol commands, documented here: http://chemapps.stolaf.edu/jmol/docs/#coloratomobject.
Hi, I used your instructions to create this Tryptamine model as a going away gift to my adviser! Thank you!
https://www.shapeways.com/model/1623207/gpyout.html?li=aeTabs
Afternoon all,
Just wanted to contribute a little bit of info about our institution, as we work heavily with molecular models using 3D printing technology and might be a useful resource for those reading this article.
At the MSOE Center for BioMolecular Modeling (http://cbm.msoe.edu/) we use Jmol, Pymol, VMD, Chimera, and other molecular visualization programs to create full-color 3D printed physical models of protein and molecular structures. These materials go directly into high school and undergraduate classrooms for educational purposes – including through our grant-funded “lending library”, where educators can borrow our models free of charge.
We have also created physical models using X-ray and MRI data, including hips, brains and other macroscopic objects.
Happy modeling everyone!
-Mark Hoelzer
MSOE Center for BioMolecular Modeling