In the late 1980s, as I recollected here[1] the equipment needed for real time molecular visualisation as it became known as was still expensive, requiring custom systems such as Evans and Sutherland PS390 workstations. One major breakthrough in making such techniques generally available on less specialised equipment was achieved by Roger Sayle[2], then working at Imperial College around 1990 and using a Silicon Graphics workstation. He greatly optimised up the rendering algorithms by creating a program called RasMol (after his initials), which meant such visualisations could very rapidly also be achieved even on a personal computer. Moving from vector display technology (the PS390) to Raster/bitmap graphics had allowed spacefilling representations of molecules containing 100s if not 1000s of atoms – and in turn enabled the new World-Wide Web to exploit the technique.[3]
Whilst Rasmol is very much still around, it also provided an inspiration for successor programs such as Jmol (based on Java) and JSMol (based on the Javascript language built into all modern web browsers). There are now many articles in the literature describing this program. In 2008 the very first post on this blog described how run it in a WordPress instance[4].
Now a new milestone in molecular visualisation has been reached – the ability to display 3 million atoms! Bob Hanson has just released Jmol/JSmol 16.1.51 which supports the BinaryCIF file format. An example of the power of both program and this new format is illustrated with the protein 8glv[5] which contains 3 million atoms (the bcif file itself is only 47.4 Mb).
The Jmol/JSmol script to load it is:
t = now();
set autobond false;
load =8glv.bcif filter "*.CA";
spacefill on;
color chain;
print now(t);and the actual rendering takes just 10-20 seconds. You can see from the screenshots below‡ that when it is zoomed in, it really does show individual atoms! Who knows what the practical atom limit is, but it is almost certainly more than three million! And it may even be possible on a mobile phone!
‡OK, you are asking why I have not loaded 8glv into this page? Well, I need to update JSmol on this site first and have encountered an issue that needs fixing.
Postcript. We are getting there. Below is a screen capture of this protein using an iPhone 15 and any of Safari, FireFox, Chrome or Edge browsers via this link (ca 12 seconds on iPhone, ~30 seconds on a 2019 iMac, ~12 seconds on an M1 Mac Studio). It even spins reasonably smoothly!
References
- H. Rzepa, "Computers 1967-2011: a personal perspective. Part 2. 1985-1989.", 2011. https://doi.org/10.59350/g4j62-4xk50
- R. Sayle, "RASMOL: biomolecular graphics for all", Trends in Biochemical Sciences, vol. 20, pp. 374-376, 1995. https://doi.org/10.1016/s0968-0004(00)89080-5
- H.S. Rzepa, B.J. Whitaker, and M.J. Winter, "Chemical applications of the World-Wide-Web system", Journal of the Chemical Society, Chemical Communications, pp. 1907, 1994. https://doi.org/10.1039/c39940001907
- H. Rzepa, "Jmol and WordPress: Loading 3D molecular models, molecular isosurfaces and molecular vibrations into a blog", 2008. https://doi.org/10.59350/pq7ds-gqr71
- T. Walton, and A. Brown, "96-nm repeat unit of doublet microtubules from Chlamydomonas reinhardtii flagella", 2023. https://doi.org/10.2210/pdb8glv/pdb

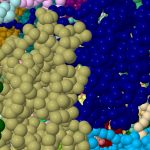
I should point out that the command shown above; load =8glv.bcif filter “*.CA”; means that 3 million atoms are not actually loaded. Its actually 503237 after filtering. Without this filter, the molecule will not in fact load into Jmol (or JSmol). But half a million atoms is still pretty good! It would be good to establish whether loading can be pushed to anything higher than 503237?