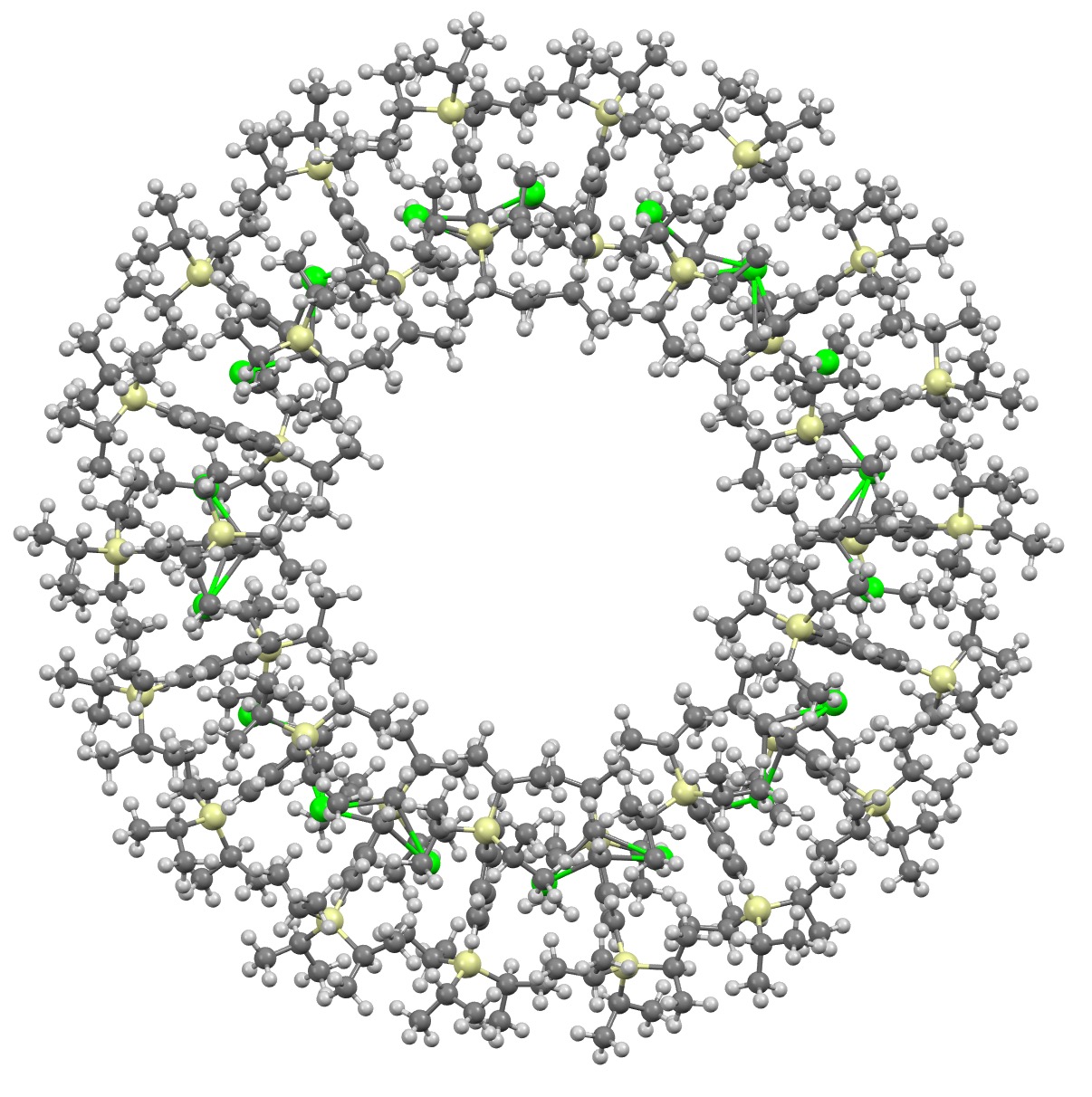
I will approach this example of a molecule-of-the-year candidate – in fact the eventual winner in the reader poll – from the point of view of data. Its a metallocene arranged in the form of a ring comprising 18 sub-units.[1] Big enough to deserve a 3D model rather than the static images you almost invariably get in journals (and C&EN). So how does one go to the journal and acquire the coordinates for such a model?
Well, nowadays most reputable journals include a “data availability” statement, which in this case is indicated using a URL-style identifier for supporting information. This means by the way that this identifier may not be persistent, since the path to the document in the string https://static-content.springer.com/esm/art%3A10.1038%2Fs41586-023-06192-4/MediaObjects/41586_2023_6192_MOESM1_ESM.pdf may change in the future according to the publishers production workflows. The Acrobat file contains the required coordinates, of which I give a small sample here:
18‐ring ‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐ 1386 Energy = ‐29312.63737385 dispersion contribution = ‐2.415738946 C 5.1700172 1.6243489 ‐11.0779621 C 5.6857216 1.5855492 ‐12.4187559 C 6.0496599 0.6048512 ‐13.3969079 C 6.1219344 ‐0.8254711 ‐13.5066237
I selected the molecule coordinates within the PDF, pasted into a text editor and then spent a few minutes removing the resulting extraneous blank lines due to the page breaks present in the PDF document (a paginated document format is NOT a good vehicle for data!). I then added further lines (topped and tailed it) to eg make it viewable using a molecular editor such as Gaussview, only to get the following error.
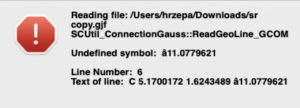
A bit of research leads to eg the following page: The difference between a dash and a minus sign. There you find four different glyphs any of which could look like a minus sign – there could in fact be more. Next, using the following resource: https://www.fontspace.com/unicode/analyzer#e=4oCQ tells us that the “-” found in the supporting information is in fact a “hyphen“. Typed from a keyboard as a “-” one learns this is a “hyphen-minus“. There is also “−” which emerges as a “Minus sign“, whilst a “–” emerges as an “EN Dash“. Confused yet? Well, it all does rather depend on whether the creator of the molecular viewing program you are about to use has included all these variations in their program code. In this case clearly not, since a hyphen is not recognised. Once you get to this stage, around 30 minutes of occasional head scratching have elapsed, and you further have figured out how to do a global find and replace of a hyphen by a minus using your preferred software.
What does all this have to do with FAIR? This means Findable, Accessible, Interoperable and Reusable. And those actions have to be possible not only by a human but by an autonomous and probably unsupervised system gathering data for machine learning or artificial intelligence. The Finding was facilitated by the “data availability” statement using the article DOI (a fully persistent identifier), but probably only a human could actually cope with the diversity of presentations for data found across multiple publishers (thus, to be technical, the access location of supporting data is rarely if ever actually declared in the metadata record associated with the DOI, which is what a machine would need to access the data). The Access in this case means resolving the URL above, but only if it does not change in the future! But the next bit, the Interoperability, is more of a challenge. Like myself, many a human might also take 30 minutes, or indeed just give up, in coping with the challenge of recognising that a hyphen is not a minus! So although we are grateful for that “data availability” statement, I dream of the day when that will in fact become a “FAIR data availability” statement!‡ Not many signs of that happening yet. I guess the AI-algorithms will in fact get smarter faster than people for coping with such issues.
Anyway, you now have a 3D model of the 18-metallocene as this year’s selected molecule of the year! Click on the image above to load it.
‡For example, the data for this post is available at a FAIR repository, with the persistent DOI identifier: https://doi.org/10.14469/hpc/13536. This contains the optimised coordinates using the PM7 method. These are very little different from the coordinates from the article, which were obtained using the PBE0/Def2-TZVP method, a remarkable calculation given it uses 21618 basis functions!
References
- L. Münzfeld, S. Gillhuber, A. Hauser, S. Lebedkin, P. Hädinger, N.D. Knöfel, C. Zovko, M.T. Gamer, F. Weigend, M.M. Kappes, and P.W. Roesky, "Synthesis and properties of cyclic sandwich compounds", Nature, vol. 620, pp. 92-96, 2023. https://doi.org/10.1038/s41586-023-06192-4
I agree, this is indeed a bit inconvenient to provide the coordinates data in the ESI PDF file (especially when you’re working with big structures that spread over several pages, and if the file is numbered then you have to delete all those page numbers etc..). A separate archive containing all the XYZ files would certainly make life much easier. The issue with the hyphen in the data, as you’ve mentioned, is quite intriguing (I’ve never encountered this before, maybe because I do not use Gaussview). By the way, Chemcraft was able to plot the structure without requiring any modifications to the text copied from the PDF.
Its a bit more than just providing XYZ coordinates. For example the provenance of these coordinates, which in this example come from a PBE0/def2‐TZVP/ECP/D4 calculation. This sort of information is best expressed as formal metadata, which has a well defined and declared structure that eg a machine could parse and hence understand. In this example, the provenance is at least declared as unstructured free text, which a human can cope with pretty well but where a machine would have to work hard to understand to the point of exact replication. I for one would be interested in more detail than this simple declaration; as I noted above, this implies a calculation was performed using 21618 basis functions, and elsewhere we find this was done using Turbomole. That is an impressive number of basis functions (I have never exceeded about 3000 in my own work) and so this program must have some amazing capabilities to do this. A triple-ζ calculation on a system with 1386 atoms is certainly something to explore!
It’s fascinating to delve into the intricacies of data availability and the hurdles that come with it, especially in scientific research. The example of the 18-ring Strontium Metallocene highlights just how crucial it is for data to be FAIR—Findable, Accessible, Interoperable, and Reusable. The effort required to convert a hyphen to a minus sign in molecular coordinates may seem trivial, but it underscores a significant issue in data interoperability. For those involved in data-intensive fields, this serves as a reminder of the importance of robust data standards and the need for continued advancements in making scientific data truly FAIR. SEI