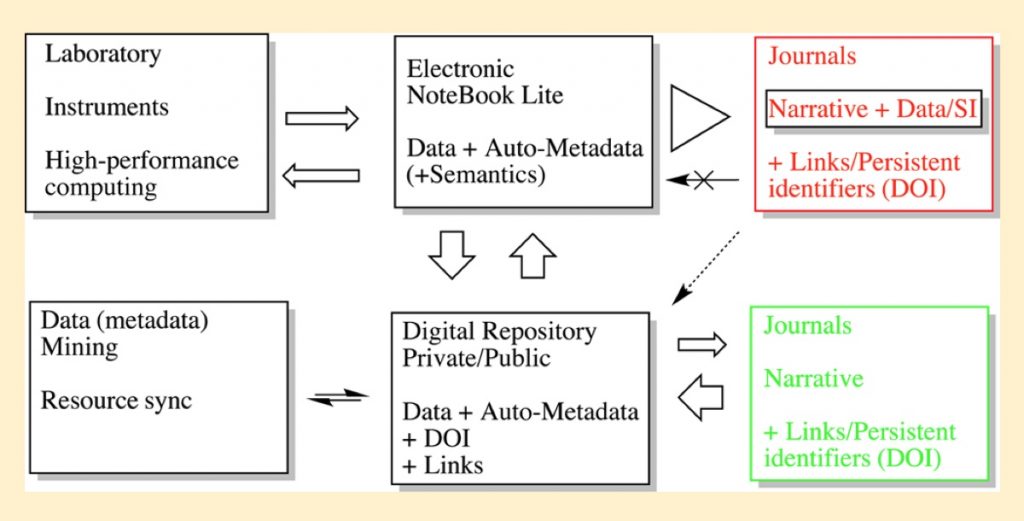
Way back in the late 1980s or so, research groups in chemistry started to replace the filing of their paper-based research data by storing it in an easily retrievable digital form. This required a computer database and initially these were accessible only on specific dedicated computers in the laboratory. These gradually changed from the 1990s onwards into being accessible online, so that more than one person could use them in different locations. At least where I worked, the infrastructures‡ to set up such databases were mostly not then available as part of the standard research provisions and so had to be installed and maintained by the group itself. The database software took many different forms and it was not uncommon for each group in a department to come up with a different solution that suited its needs best. The result was a proliferation of largely non-interoperable solutions which did not communicate with each other. Each database had to be searched locally and there could be ten or more such resources in a department. The knowledge of how the system operated also often resided in just one person, which tended to evaporate when this guru left the group.
After the millennium, two newcomers started to appear, one being called an ELN (electronic laboratory notebook) and the second a data repository. The first was a heavily customised database containing research data as obtained from instruments, computers, images/video, chemical structure drawings etc. ELNs, even to this day, have limitations of interoperability with other ELNs and the contents of an ELN are often closed, requiring authentication credentials to access. The data repository also started to appear in chemistry around this period. Even in its early incarnations, it could be associated with an ELN “front end” as part of the data pipeline; an early example of this coupling is described here.[1] Another key phrase that became associated with repositories starting around 2014 was the concept of FAIR, including ideas such as the Findability (discoverability) and Interoperablity of data,† a theme often explored and illustrated on this blog.
These last seventeen years has seen organisations such as funding agencies and publishers increasingly mandating the use of such data management methods, using either a repository on its own or a combination of an ELN and repository as routine operations in research activity and publication processes. The close coupling of an ELN and repository is still however uncommon.
A colleague recently alerted me to a computational chemistry repository first launched in 2014; www.iochem-bd.org Reading the about text, I found these statements;
- Chem-BD is a digital repository aimed to manage and store Computational Chemistry files.
- Goals: Build a distributed database of computational chemistry results: reduce size and increase value.
- Set a common data standard among all quantum chemistry legacy formats (XML – CML[2])
So this is both a database and a data repository, as well as espousing a commendable common data standard![2] I decided to explore the first two aspects here using this resource as an example.
- Whilst the absolute distinction between the two types can be blurry, the crucial difference between the two is that a database functions on curation via a structured index of the data, whilst a repository aspires to having FAIR attributes primarily through its metadata as exposed by registration (metadata is data describing the data).
- A database holds this data index locally and the Findability of the data is associated purely with the functionality of the database. The data structures are defined by a database schema, describing in detail all the terms indexed (a key and its value) and searched using the values of these key pairs. This schema is unlikely to be exactly the same as e.g. databases on related topics, largely because the database is self-contained and self-consistent.
- A data repository also uses a schema (DOI: 10.14454/3w3z-sa82 and[3])♠ to express the key pairs, but this time it is expressed as metadata. Now, this metadata is registered externally to the repository using a registration agency.[3] The metadata for each deposited object is assigned a persistent identifier known as a DOI. Although it might be indexed and searchable locally, it must be capable of also being searched in aggregated/federated form using services provided by registration or other agencies. This independence of metadata is part of those FAIR criteria.
- Whereas a database can be very finely grained in order to describe individual properties of an object, repository metadata tends to be more coarsely grained to describe the object as a whole, to place it in context and to impart provenance.
- Both databases and repositories can have what is called an API (application programmer interface) to allow machine access (the A of FAIR) to the contents. Accessing the former would normally require bespoke code to be written and possibly authentication credentials, whereas information to access to repository held data is provided via the registered metadata (which does not normally require credentials). Access to the repository may also require code, but if the metadata is carefully standardised by adherence to the schema, the code can be made more general than that required for a database.♥
- A typical entry in the www.iochem-bd.org repository has a DOI of 10.19061/iochem-bd-4-36
- This DOI is registered with the CrossRef agency, one normally used for registering journal articles, rather than DataCite which is used for registering data and other research objects. The metadata for this DOI can be viewed using the resolution service https://api.crossref.org/works/10.19061/iochem-bd-4-36/transform/application/vnd.crossref.unixsd+xml and shows that it largely contains the bibliographic information typical of a journal article. So in this sense it is certainly a repository, but using a metadata schema that is more frequently used for journal articles than for data sets.
- The CrossRef metadata record also has an item <resource>https://www.iochem-bd.org/handle/10/235025</resource> which points to the so-called landing page for that item, but information about the properties of the actual data itself must be instead obtained directly from the repository.
- Because the metadata describing the data is only held at this repository and not elsewhere (a local metadata record), it can only be queried locally and the query cannot be upon aggregated metadata provided by the registration agency. A machine query would have to be constructed by coding a suitable request using the API provided for the database aspect of this repository.
This example has served to highlight just a few of the often quite subtle distinctions between eg a database and a data repository and that some examples can indeed be both. It also highlights that repositories can have the attributes of FAIR, which in themselves are driven by asking “what could a machine do to obtain data?”♥ rather than what could a human achieve by browsing. So another question that arises when evaluating the characteristics of a repository is whether each item held there has a FAIR-enabling metadata record describing the data, a record which is registered in a manner that can be aggregated and hence used to find and access content across multiple independent repositories.
This post has DOI 10.14469/hpc/10043
‡Indeed in that era, few online/Internet infrastructures were available as part of departmental resources. See also here. †In this last regard, I note a workshop devoted largely to such interoperability and machine access in chemistry coming up soon; https://www.cecam.org/workshop-details/1165 ♠The CrossRef schema is not referenced using an assigned DOI: data.crossref.org/reports/help/schema_doc/5.3.1/.♥An example can be seen at doi: 10.14469/hpc/10059 Here, invoking a hyperlink based purely on the data DOI and the data media type required in turn calls code (Javascript) which retrieves the metadata held for that DOI and parses it to identify whether it indicates the presence of a file manifest. If it does, it identifies the type of manifest (ORE in this case) and the media types the manifest points to and finally uses that manifest to then retrieve data filtered by media type and pipes it into a visualiser (JSmol). In this case the endpoint is visualisation, but it could also be eg piped into an AI/ML program for analysis. In this case only one instance of data is machine retrieved, but in principle it could be a multitude of data files obtained from a multitude of different locations and based on a multitude of criteria as filtered by suitable searches of registered metadata.[4]
References
- M.J. Harvey, N.J. Mason, and H.S. Rzepa, "Digital Data Repositories in Chemistry and Their Integration with Journals and Electronic Notebooks", Journal of Chemical Information and Modeling, vol. 54, pp. 2627-2635, 2014. https://doi.org/10.1021/ci500302p
- P. Murray-Rust, and H.S. Rzepa, "Chemical Markup, XML, and the Worldwide Web. 1. Basic Principles", Journal of Chemical Information and Computer Sciences, vol. 39, pp. 928-942, 1999. https://doi.org/10.1021/ci990052b
- H. Cousijn, T. Habermann, E. Krznarich, and A. Meadows, "Beyond data: Sharing related research outputs to make data reusable", Learned Publishing, vol. 35, pp. 75-80, 2022. https://doi.org/10.1002/leap.1429
- H.S. Rzepa, and S. Kuhn, "A data‐oriented approach to making new molecules as a student experiment: artificial intelligence‐enabling FAIR publication of NMR data for organic esters", Magnetic Resonance in Chemistry, vol. 60, pp. 93-103, 2021. https://doi.org/10.1002/mrc.5186