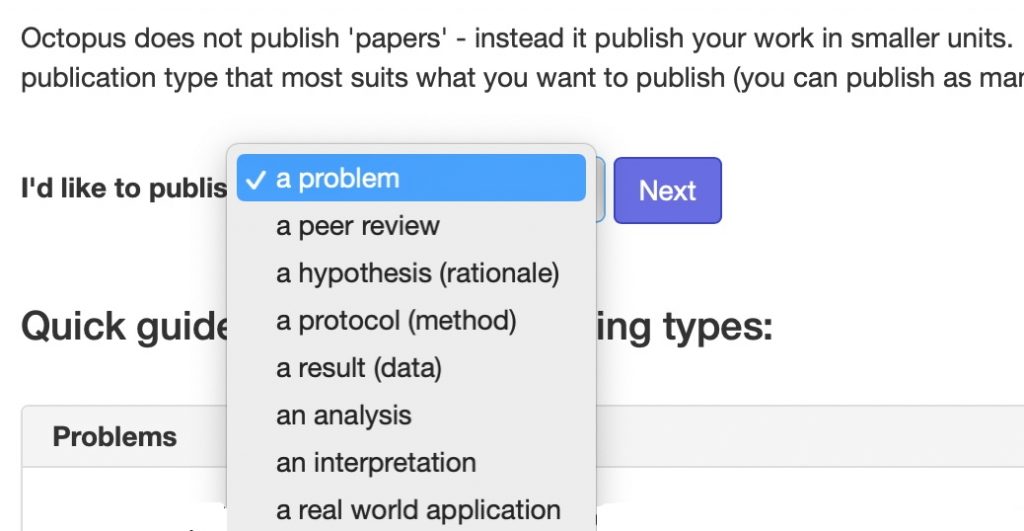
In 2011, I suggested that the standard monolith that is the conventional scientific article could be broken down into two separate, but interlinked components, being the story or narrative of the article and the data on which the story is based. Later in 2018 the bibliography in the form of open citations were added as a distinct third component.[1] Here I discuss an approach that has taken this even further, breaking the article down into as many as eight components and described as “Octopus publishing” for obvious reasons. These are;
- The problem being addressed
- An original hypothesis/theoretical Rationale for the problem
- A method or protocol for testing the hypothesis in the form of experiments or modelling
- The data resulting from these experiments
- Analysis of this data
- Interpretation of the analysis in terms of the original hypothesis
- Translation/application to a real world problem (an extrapolation if you like of the original problem)
- A review of any of the previous seven items.
Items 1-3, 5-6 and probably 7 are the dis-assembled components of the standard concept of a scientific publication and item 4 is the data component I refer to in my introduction above. Interestingly, the article bibliography is not separated out into these components, and is presumably distributed throughout the resulting fragments.♥ The essential concept behind this dis-assembly is that each component can rest on its own, provided it is contextually and bi-directionally linked into the others. The author(s) of any individual component will get credit/recognition for that component. A conventional mapping would be that the same author set would be responsible for all the individual items, whilst recognising that each component could in fact have its separate authorship. Thus one might get credit for just suggesting a problem, or for suggesting a protocol for its testing, for acquiring just the data, or for proposing an original analysis or interpretation. Any author’s reputation would then be established by the integrated whole of their contributions across the whole range of article components (leaving aside how the weightings of each individual contribution would be decided).†
I decided to try to give it a go via a prototype at https://science-octopus.org/publish which as you can see is one of the eight above. The process starts with the prospective author providing their ORCID credentials, against which unique metadata is presumably generated.
The next stage however is more interesting; Unlike any other publication platform, Octopus requires all publications to be linked to others that already exist. At the current stage of development, the prototype only has a few entries linked to COVID-19 as a topic, which I did not feel able to add to. Selecting one of those at random, one is asked to link any associated data via a DOI, then the optional indication of appropriate keywords, followed by standard questions about funding sources, conflicts of interest and license declaration. I wanted to use CC0, but this was not an option.
Finally at this stage a question about any other related publications, with the list known to Octopus being offered as suggestions. Next came the main opportunity to insert prose related to the information already provided (this part constituting the conventional article component). This is also the only opportunity to add a bibliography, and the citations would be part of that document, albeit not identified as a citation for the inner workings of the Octopus system. Then comes a Publish now button. No persistent identifier (DOI) is generated in this prototype system, but will be in the production system. A final screen that has four options, including to write a review (of one’s own work?) or to Red Flag the item. The latter can eg arise from plagiarism or any other expression of concern.
Many questions arise about this new approach and I will only note only three. One relates to “Why would I want to publish in Octopus” in the FAQ section. I quote “The traditional system is not only slow and expensive, but the concept of “papers” is not a good way of disseminating scientific work in the 21st century and “Publish it now and establish priority – once it”s out in Octopus it”s yours“. There are many experiments that strive to address this generic issue that the conventional research paper is no longer entirely fit for purpose. One I can supply here is that of “Preprint servers” such as ChemRxiv where one significant motivation is also to “establish priority”.
An aspect of interest to myself is how the metadata for all these eight components will be expressed. Presumably when mature, all eight components will have their own DOI or persistent identifier (PID) and hence all eight will also have a metadata record.♥ These records will formally establish the relationships between the components, and ultimately could be used to construct a PID Graph not only between those components but to the rest of the PID “multiverse” of articles, data, citations and other research objects. Will Octopus join in this PID graph world?
And finally, the greatest challenge to a new paradigm such as Octopus is how quickly will the existing established culture of “publish a blockbuster article in one of the top ten (chemistry) journals” to establish your career evolve into the dis-assembled approach described here? It has taken preprint servers the best part of 25 years to really get going in e.g. chemistry where there are now around 9600 preprints on a variety of topics.‡ I suspect some subject disciplines may be harder to crack than others (and chemistry may well be amongst these!).
† Around 2009 or so I ran a student experiment using a Wiki which had some aspects of this approach. Students were asked to do a project on the topic of either a molecule from a suggested list of around 30, or a molecule entirely of their own choosing. The students spontaneously split themselves into three groups. The first were students who wrote the story entirely by themselves, submitted it for credit and did not welcome others as co-authors. The second (largest) group where those that contributed to multiple topics, very much in the manner of Wikipedia itself. Their credit was the sum of their contributions. The final group chose not to tell a story about a molecule, but to help everyone else with the infrastructure of doing so (the protocol if you like) by writing templates which simplified authoring, or correcting errors in existing stories etc.
‡Arguably a measure of the impact of these 9600 preprints is how many of them have eventually appeared in fully peer reviewed form in journals. That statistic may not be known. Also of interest would be some analysis for those that did end up in journals of how they evolved between eg V1 of the preprint and the final “version of record”.
♥Metadata is structured according to a specified schema. Currently, journal publishers use the CrossRef schema for this purpose, which contains a full description of e.g. the bibliography of an article. Data publishers use the DataCite Schema, which has less focus on bibliography. It will be of interest to see how such schemas are applied to the eight components of an Octopus scientific record.
References
- D. Shotton, "Funders should mandate open citations", Nature, vol. 553, pp. 129-129, 2018. https://doi.org/10.1038/d41586-018-00104-7