As the Internet and its Web-components age, so early pages start to decay as technology moves on. A few posts ago, I talked about the maintenance of a relatively simple page first hosted some 21 years ago. In my notes on the curation, I wrote the phrase “Less successful was the attempt to include buttons which could be used to annotate the structures with highlights. These buttons no longer work and will have to be entirely replaced in the future at some stage.” Well, that time has now come, for a rather more crucial page associated with a journal article published more recently in 2009.[1]
The story started a few days ago when I was contacted by the learned society publisher of that article, noting they were “just checking our updated HTML view and wanted to test some of our old exceptions“. I should perhaps explain what this refers to. The standard journal production procedures involve receiving a Word document from authors and turning that into XML markup for the internal production processes. For some years now, I have found such passive (i.e. printable only) Word content unsatisfactory for expressing what is now called FAIR (Findable, accessible, inter-operable and re-usable) data. Instead, I would create another XML expression (using HTML), which I described as Interactive Tables and then ask the publisher to host it and add that as a further link to the final published article. I have found that learned society publishers have not been unwilling to create an “exception” to their standard production workflows (the purely commercial publishers rather less so!). That exceptional link is http://www.rsc.org/suppdata/cp/b8/b810301a/Table/Table1.html but it has now “fallen foul of the java deprecation“.
Back in 2008 when the table was first created, I used the Java-based Jmol program to add the interactive component. That page, when loaded, now responds with the message:
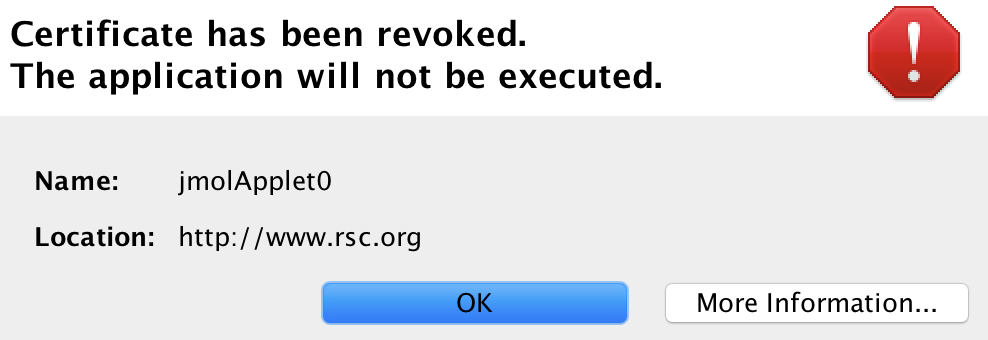
This I must emphasise is nothing to do with the publisher, it is the Jmol certificate that has been revoked. That of itself requires explanation. Java is a powerful language which needs to be “sandboxed” to ensure system safety. But commands can be created which can access local file stores and write files out there (including potentially dangerous ones). So it started to become the practise to sign the Java code with the developer certificate to ensure provenance for the code. These certificates are time-expired and around 2015 the time came to renew it. Normally, when such a certificate is renewed, the old one is allowed to continue operation. On this occasion the agency renewing the certificate did not do this but revoked the old one instead (Certificate has been revoked, reason: CESSATION_OF_OPERATION, revocation date: Thu Oct 15 23:11:18 BST 2015). So all instances of Jmol with the old certificate now give the above error message.
The solution in this case is easy; the old Jmol code (as JmolAppletSigned.jar) is simply replaced with the new version for which the certificate is again valid. But simply doing that alone would merely have postponed the problem; Java is now indeed deprecated for many publishers, which is a warning that it will be prohibited at some stage in the future.‡ So time to bite the bullet and remove the dependency on Java-Jmol, replacing it with JSmol which uses only JavaScript.
Changing published content is in general not allowed; one instead must publish a corrigendum. But in this instance, it is not the content that needs changing but the style of its presentation (following the principle of the Web of a clear-cut separation of style and content). So I set out to update the style of presentation, but I was keen to document the procedures used. I did this by commenting out non-functional parts of the style components of my original HTML document (as <!– comment –>) and adding new ones. I describe the changes I made below.
- The old HTML contained the following initialisation code: jmolInitialize(".","JmolAppletSigned.jar");jmolSetLogLevel('0'); which was commented out.
- New scripts to initialize instead JSmol were added, such as:
<script src="JSmol.min.js" type="text/javascript"> </script> - I added further scripts to set up controls to add interactivity.

- The now deprecated buttons had been invoked using a Jmol instance: jmolButton('load "7-c2-h-020.jvxl";isosurface "" opaque; zoom 120;',"rho(r) H")
- which was replaced by the JSmol equivalent, but this time to produce a hyperlink rather than a button (to allow the greek ρ to appear, which it could not on a button): <a href="javascript:show_jmol_window();Jmol.script(jmolApplet0,'load 7-c2-020.jvxl;isosurface "" translucent;spin 3;')">ρ(r)</a>,
- Some more changes were made to another component of the table, the links to the data repository. Originally, these quoted a form of persistent identifier known as a Handle; 10042/to-800. Since the data was deposited in 2008, the data repository has licensed further functionality to add DataCite DOIs to each entry. For this entry, 10.14469/ch/775. Why? Well, the original Handle registration had very little (chemically) useful registered metadata, whereas DataCite allows far richer content. So an extra column was added to the table to indicate these alternate identifiers for the data.
- We are now at the stage of preparing to replace the Java applet at the publishers site with the Javascript version, along with the amended HTML file. The above link, as I write this post, still invokes the old Java, but hopefully it will shortly change to function again as a fully interactive table.
- I should say that the whole process, including finding a solution and implementing it took 3-4 hours work, of which the major part was the analysis rather than its implementation.
It might be interesting to speculate how long the curated table will last before it too needs further curation. There are some specifics in the files which might be a cause for worry, namely the so-called JVXL isosurfaces which are displayed. These are currently only supported by Jmol/JSmol. They were originally deployed because iso-surfaces tend to be quite large datafiles and JVXL used a remarkably efficient compression algorithm (“marching cubes”) which reduces the cube size one hundred-fold or more. Should JSmol itself become non-operational at some time in the (hopefully) far future (which we take to be ~10 years!) then a replacement for the display of JVXL will need to be found. But the chances are that the table itself will decay “gracefully”, with the HTML components likely to outlive most of the other features. The data repository quoted above has itself now been available for ~12 years and it too is expected to survive in some form for perhaps another 10. Beyond that period, no-one really knows what will still remain.
You may well ask why the traditional journal model of using paper to print articles and which has survived some 350 years now, is being replaced by one which struggles to survive 10 years without expensive curation. Obviously, a 3D interactive display is not possible on paper.[2] But one also hears that publishers are increasingly dropping printed versions entirely. One presumes that the XML content will be assiduously preserved, but re-working (transforming, as in XSLT) any particular flavour of XML into another publishers systems is also likely to be expensive. Perhaps in the future the preservation of 100% of all currently published journals will indeed become too expensive and we might see some of the less important ones vanishing for ever?†
‡Nowadays it is necessary to configure your system or Web browser to allow even signed valid Java applets to operate. Thus in the Safari browser (which still allows Java to operate, other popular browsers such as Chrome and Firefox have recently removed this ability), one has to go to preferences/security/plugin-settings/Java, enter the URL of the site hosting the applet and set it to either “ask” (when a prompt will always appear asking if you want to accept the applet) or “on” when it will always do so. How much longer this option will remain in this browser is uncertain.
†In the area of chemistry, an early pioneer was the Internet Journal of Chemistry, where the presentation of the content took full advantage of Web-technologies and was on-line only. It no longer operates and the articles it hosted are gone.
References
- H.S. Rzepa, "Wormholes in chemical space connecting torus knot and torus link π-electron density topologies", Phys. Chem. Chem. Phys., vol. 11, pp. 1340-1345, 2009. https://doi.org/10.1039/b810301a
- H.S. Rzepa, "Transclusions of data into articles", 2013. https://doi.org/10.6084/m9.figshare.797481
Tags: Applet, compression algorithm, computing, Cross-platform software, HTML, HTML element, Internet Journal, Java, Java applet, Java platform, jmol, Markup languages, Open formats, publishers site, publishers systems, technology moves, Technology/Internet, the Internet Journal, Web browser, web technologies, Web-components age, XML, XSLT
If you try the link
http://www.rsc.org/suppdata/cp/b8/b810301a/Table/Table1.html
today, you will find no trace of Java (or any error messages such as the one above). Thanks to the RSC for helping this curation.
“They were originally deployed because iso-surfaces tend to be quite large datafiles and JVXL used a remarkably efficient compression algorithm (“marching cubes”) which reduces their size ten-fold or more.”
For potential interest, I am currently working on a Python tool (https://github.com/bskinn/h5cube) and accompanying file specification (current draft at http://h5cube-spec.readthedocs.io/en/draft/index.html) for the conversion of volumetric data in the Gaussian CUBE format into a compressed form using the HDF5 data format (https://support.hdfgroup.org/HDF5/). HDF5 incorporates compression algorithms that are especially powerful for data such as visualization isosurfaces, where semi-lossy compression (truncation of precision and thresholding of retained data) is acceptable.
For example, the following is an 0.002 e-/B^3 isosurface of the electron density of benzene with a 250x250x250 voxel grid, truncated to three digits of precision past the decimal and thresholded to between 0.012 and 0.00033 (VMD v1.9.1):
The high density of the voxel grid can be discerned from the wireframe-rendered isosurface on the right. The CUBE file used to generate this image was 198MB; it was decompressed from an “h5cube” file that is 2.5MB (MB, not GB) in size (~79-fold compression). While this compressed data file is no longer suitable for volumetric analysis (e.g., QTAIM) due to truncation, nor for rendering of isosurfaces far from the target 0.002 value due to thresholding, it is more than sufficient to provide an attractive visualization. The use of CUBE data generated on a smaller voxel grid and compressed with stricter truncation and/or thresholding would provide an even more compact h5cube file.
Even more dramatic compression is possible for “binary” volumetric data such as QTAIM and ELF basins, where each voxel is either “in” or “out” of the relevant volume. The following is the electron density basin for one of the hydrogen atoms of benzene, generated by Multiwfn v3.3.7 with a 204x211x149 voxel grid, truncated to zero digits of precision past the decimal (VMD v1.9.1):
The CUBE file used for this image was 82MB; it was decompressed from an h5cube file 156kB in size (~530-fold compression). To note, the granularity of the boundary is an inevitable artifact of the binary in/out nature of the volumetric dataset, and is not due to the h5cube compression.
Some preliminary compression-ratio benchmarking for the h5cube file format can be found at the Google Sheet here: https://docs.google.com/spreadsheets/d/1AajEYpacgq48X72_HuarVLVA517ZY4htaEWf1tLY5Cc/edit#gid=0
Unfortunately, the Python tool is not yet really in production condition, and the specification is, as noted, still in draft form. Further, AFAIK no visualization software is able to read these compressed files directly at this time, though there is no technological impediment to modifying them to do so. Once I get the tool and specification to a reasonably “stable” condition, hopefully I can begin to make a convincing case for the incorporation of the format as at least a readable option in one or more software packages.
My goal for this project is to revolutionize the sharing of volumetric data outputs from computational chemistry calculations. I believe the h5cube format has the potential to be fully FAIR, though a significant extension of the file specification (into various metadata) beyond just what is present in a normal CUBE file would almost certainly be required. Feedback/suggestions are most welcome!
Here are some more FAIR data curations with the same publisher:
1. http://www.rsc.org/suppdata/cp/b9/b911817a/Table/Table1.html
2. http://www.rsc.org/suppdata/CC/c0/c0cc04023a/Table1/index.html
3. http://www.rsc.org/suppdata/cc/c2/c2cc33676f/dyotropic/index.html
4. http://www.rsc.org/suppdata/cc/c3/C3CC46720A/C3CC46720A/index.html
5. http://www.rsc.org/suppdata/dt/b8/b810147g/Table1.html
6. http://www.rsc.org/suppdata/cc/b9/b913295c/table/tables_index.html
The functional versions should appear shortly (post 12.06.2017).
PS. A list of early interactive figures and tables is archived at DOI: rnp.
I have some protein animations on my webasite, originally using MDL Chime, and (since a few years ago) JMol. They were still running fine in February, but I cannot open them using Chrome, Firefox or even Internet explorer. Would you mind checking whether they can still be seen using Safari?
http://homepage.ufp.pt/pedros/anim_jmol/instructen.htm
Re: the Internet Journal of Chemistry. Do you know who was in charge of that journal, and whether any external archiving (using LOCKSS or CLOCKSS) was enabled?
Yes, they still work in Safari. One has to configure the “exception site list” in the Java control panel.

For the record, I have also tried the Safari technology preview, which is a sampler of what might be in the next version of macOS, High Sierra. It works there too! We might presume that this means its life might be assured for another year or so. But I will test again when High Sierra itself is released.
Re: Internet J. Chemistry, the editor was Steve Bachrach. I am not aware of any external archiving, but Steve would have to confirm that.
One of the curated articles noted above actually illustrates two different methods of updating a Jmol page to use instead JSmol.
http://www.rsc.org/suppdata/cc/c2/c2cc33676f/dyotropic/index.html
converts the Jmol syntax to use JSmol instead. This requires a fair bit of editing. There is however an easier method:
http://www.rsc.org/suppdata/cc/c2/c2cc33676f/dyotropic/index-jmol2.html
This leaves the original Jmol code intact, but instead invokes an additional script; <script type="text/javascript" src="js/Jmol2.js"></script> to perform the syntactic conversions. You should not be able to detect any differences between the two pages in the screen display.
I chose the first option since it has one fewer long term dependency (the Jmol2.js script itself), but using this second method is certainly far easier. Both methods are fully documented at http://wiki.jmol.org/index.php/Jmol_JavaScript_Object#Conversion_using_the_Jmol2.js_.27adapter.27_library.
Thanks for checking that Safari still reads my pages. My pages were made using JMolShell (by Craig T. Martin, U. MAssachussets Amherst) and transforming them to JSMol is not as straightforward as your methods. I have been doing those transformations by hand today and I expect to upload the new JavaScript-compliant pages tomorrow. I will have to wait a few days for them to become externally accessible , but I think that now my problems have been solved. Thank you very much for telling me about JSMol.!
An organisation known as CLUE (Cooperation And Liaison Between Universities And Editors) has recently produced some Recommendations On Best Practice (for Research Data), doi: 10.1101/139170. They state that Essential research data and peer review records should be retained for at least 10 years.
As I note above, it is very likely that if that data is to be retained as FAIR (findable, accessible, inter-operble and re-usable) it may well need at least one occasion of curation over a ten year period. The above post illustrates how the data wrapper (referred to variously as an interactive table or a WEO) was curated. I did not mention that the data repository itself, which is referenced throughout these tables and WEOs, has also been curated during this period. That curation has taken two forms:
1. The software used for the repository (DSpace in an instance known as SPECTRa, doi: 10042/13) is periodically updated to the latest version, to ensure the latest features are supported and new features are added.
2. The metadata underlying the data is updated to conform to any schema changes (or interpretations) required by the metadata aggregator, DataCite. Thus we have recently updated the metadata for each item in the repository to better conform to Version 4 of the DataCite schema (it previously conforming to V3).