- Go to the article[1] via its “landing page” and there I (as a human) navigated to the supporting information. Could automated software have done this I wonder if it were not familiar with the journal?
- There I found a PDF file and two MP4 movies. I know movies are unlikely to contain FAIR data, so I try the former. On pages 16-17 you find the space group, cell dimensions and fractional atomic coordinates. Its not really formatted to be “I” (copying/pasting out of PDF can be a challenge) and you have to be familiar with what is a specialised format (neither A nor really R then) and some knowledge of appropriate crystallographic software or procedure to convert Table S1 and S2 into an inter-operable format such as CIF (crystallographic interchange format).
- The main article does have the following statement: Further details of the crystal structure investigation(s) may be obtained from the Fachinformationszentrum Karlsruhe, 76344 Eggenstein-Leopoldshafen (Germany), on quoting the depository number CSD-430054. Do they want you to write them a letter?
- Well, a bit of Googling reveals https://www.fiz-karlsruhe.de/en/leistungen/kristallographie/kristallstrukturdepot/order-form-request-for-deposited-data.html as the required online link (why could that not be shortened and included in the article?)
- This form has not quite yet caught up with modern journal practice. The form stipulates a page number is apparently mandatory, but although this article is fully published, it is too new to have one. I wrote “not assigned yet” and hoped for the best; a “clever” non-human script might always decide the data type of this response is wrong and reject the request! There is no field for the article DOI, which is really all the information that is needed. I pasted that into the “volume number” and again crossed my fingers.
- Two days later, whilst awaiting a response to the above, I revisited Table S1/S2 but now armed with a sample CIF file for the space group P 2/c and using a text editor, inserted into it the values found in these tables (~15 minutes). The result is shown below.‡
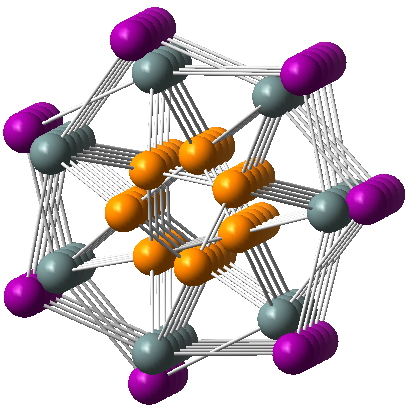
This double helix is not of the complementary type found in DNA but a concentric one. The inner helix of a chain of P atoms is enclosed by the outer helix (winding in the same sense, anticlockwise as shown above) of a Sn-I-Sn-I chain. Click on the diagram above to load the 3D coordinates and inspect this for yourself.
The article reporting this structure[1] is full of fascinating insights into this new material. Time will no doubt tell whether it has exploitable properties. Meanwhile, when the CIF file arrives from my query above, I will make it available here as properly FAIR data.
‡Use of Java and Jmol to display models is getting more and more fraught. Although it can still be made to work if you enter the host as a security exception, this is quite a deterrent. You can see a non-Java version of this post here. For reasons which have not been established, the JSmol plugin which enables this mode does not work at all on the present blog.
References
- D. Pfister, K. Schäfer, C. Ott, B. Gerke, R. Pöttgen, O. Janka, M. Baumgartner, A. Efimova, A. Hohmann, P. Schmidt, S. Venkatachalam, L. van Wüllen, U. Schürmann, L. Kienle, V. Duppel, E. Parzinger, B. Miller, J. Becker, A. Holleitner, R. Weihrich, and T. Nilges, "Inorganic Double Helices in Semiconducting SnIP", Advanced Materials, vol. 28, pp. 9783-9791, 2016. https://doi.org/10.1002/adma.201603135
Really interesting. Thanks for this fascinating post.
Four days after my request for the original data, I received the CIF file linked here. I have also edited that file to expand the unit cell to show the helix better and you can get that file linked here.