In March, I posted from the ACS meeting in San Diego on the topic of Research data: Managing spectroscopy-NMR, and noted a talk by MestreLab Research on how a tool called Mpublish in the forthcoming release of their NMR analysis software Mestrenova could help. With that release now out, the opportunity arose to test the system.
I will start by reminding that NMR data associated with a published article is (or should be) openly free: one should not need a subscription to the journal to access it (although one might in order to find it). Now, NMR data as it emerges from a spectrometer is highly sophisticated, comprising a collection of (sometimes) binary proprietary files containing the measured free induction decays (FID). Turning this raw data into an interpretable NMR spectrum, the visual form of the data that so appeals to human beings, is non trivial. This requires what may be highly sophisticated software and that in turn means that it may be a commercial product. Of course there are also examples of non-commercial open software packages that are best-of-breed; indeed in its early life-cycle MestreNova was known as MESTREC before becoming a commercial product. Could one achieve the benefits of both open and fully functional NMR data with no loss from the original instrument coupled with the ability to apply top-quality software for its analysis in an open manner? This is a demonstration of how Mpublish achieves this.
- Invoke the URL data.datacite.org/chemical/x-mnpub/10.14469/hpc/1087 from a browser
- This action queries the metadata deposited with DataCite for the doi 10.14469/hpc/1087 and retrieves the first instance of any file associated with that dataset that has the format type chemical/x-mnpub. You can directly view this metadata by invoking just data.datacite.org/10.14469/hpc/1087 where you can find both mnpub and mnova formats listed. A command such as data.datacite.org/chemical/x-mnpub/10.14469/hpc/1087 allows the file retrieval to be incorporated into automated workflows based just on the doi and the media type desired. Note my parenthetical comment above about finding data; here you only need its doi to retrieve it!
-
The URL above downloads a small text file with the suffix .mnpub which contains in essence two components:
- A URL pointing directly to an .mnova file at the repository for which the doi has been issued
- A signature key derived used to verify that the public key of the publisher (the data repository in this instance) was counter-signed by Mestrelab.
-
If you now download the application program and install it (but for the purpose of this demonstration, ignore any requests to try to license the program. Use it unlicensed) and open the .mnpub file using it, you should get the below.The application program has checked the signature key, and if valid, proceeds to download a full data file (a .mnova file in this case), and to analyze and display it within the program. The data is fully active; it can be manipulated and analysed. Notice in the picture below, the red arrow points to the state of the license, in this case not present.
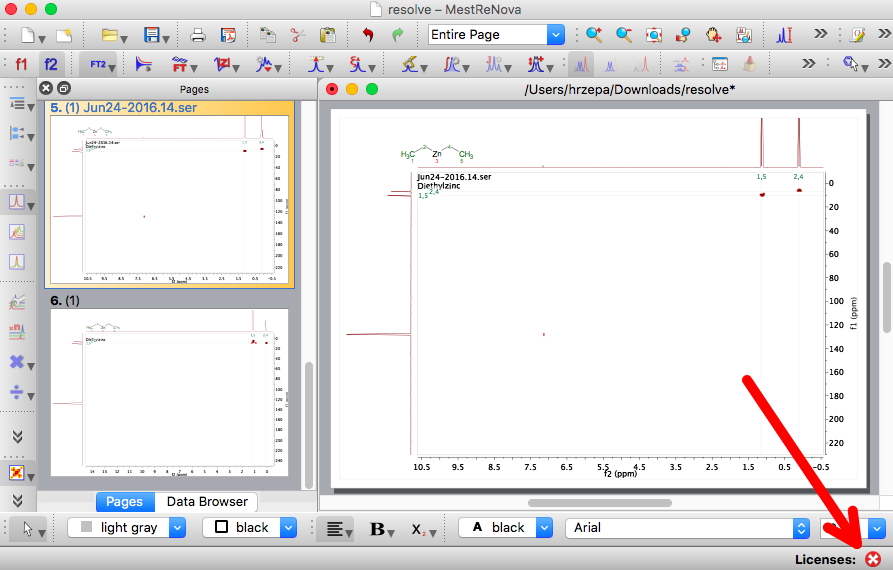
- It is also possible to apply this procedure to the raw data as it emerges from the (Bruker) spectrometer, and compressed into a .zip archive. The MestreNova software will automatically process the contents by applying various default parameters, although the result may not correspond exactly to that present in e.g. the equivalent .mnova file (which may have had specific parameters applied).
It is my hope that anyone who records NMR data and processes it using software such as MestreNova will now consider using the mechanism above to accompany their submitted articles, rather than just automatically pasting a static image of the spectrum into a PDF file as "supporting information". This is part of what is meant by "managed research data" (RDM).
One cannot help but note that many types of scientific instrument nowadays come with bespoke software for analysing the data they produce. Very often this software is unavailable to anyone who has not purchased the instrument itself. To make the data available to others, the processed data and its visual interpretation often have to be reduced, with much consequent information loss, to a lowest common denominator format such as Acrobat/PDF. Here we see a mechanism for avoiding any such information loss whilst enabling, for that dataset only, the full potential for (re)analysing the data. It will be interesting to see if other examples of this model or its equivalent emerge in the near future.
Tags: Acrobat, analysis software, chemical, Chemistry, City: San Diego, format type chemical/x-mnpub, media type, Mestrenova, non-commercial open software packages, Nuclear magnetic resonance, Nuclear magnetic resonance spectra database, Nuclear magnetic resonance spectroscopy, PDF, public key, Science, Scientific method, spectroscopy, Technology/Internet
The procedure described above works for downloading .mnova files, but a bug prevented the same procedure working for Bruker .zip archives. The release of MestreNova 11.0.1 fixes this bug. If you try
http://data.datacite.org/chemical/x-mnpub/10.14469/hpc/1095 this will now retrieve the .zip archive when you open the downloaded .mnpub file into MestreNova.
Some real-world examples of the technique described above are starting to appear.
1. Epimeric Face-Selective Oxidations and Diastereodivergent Transannular Oxonium Ion Formation-Fragmentations: Computational Modelling and Total Syntheses of 12-Epoxyobtusallene IV, 12-Epoxyobtusallene II, Obtusallene X, Marilzabicycloallene C and Marilzabicycloallene D, J. Org. Chem., 2016, doi: 10.1021/acs.joc.6b02008 with data at doi: 10.14469/hpc/1267
2. Addition of Carbon–Fluorine Bonds to a Mg(I)–Mg(I) Bond: An Equivalent of Grignard Formation in Solution, J. Am. Chem. Soc., 2016, doi: 10.1021/jacs.6b08104 with data at doi: 10.14469/hpc/1726
3. Chemoselective Polymerizations from Mixtures of Epoxide, Lactone, Anhydride, and Carbon Dioxide, J. Am. Chem. Soc., 2016, doi: 10.1021/jacs.5b13070 with data at doi: 10.14469/hpc/272
This collection DOI: 10.14469/hpc/1053 summarises our implementation of the Mpublish project. It includes an article recently published which goes into a little more detail about the implementation.