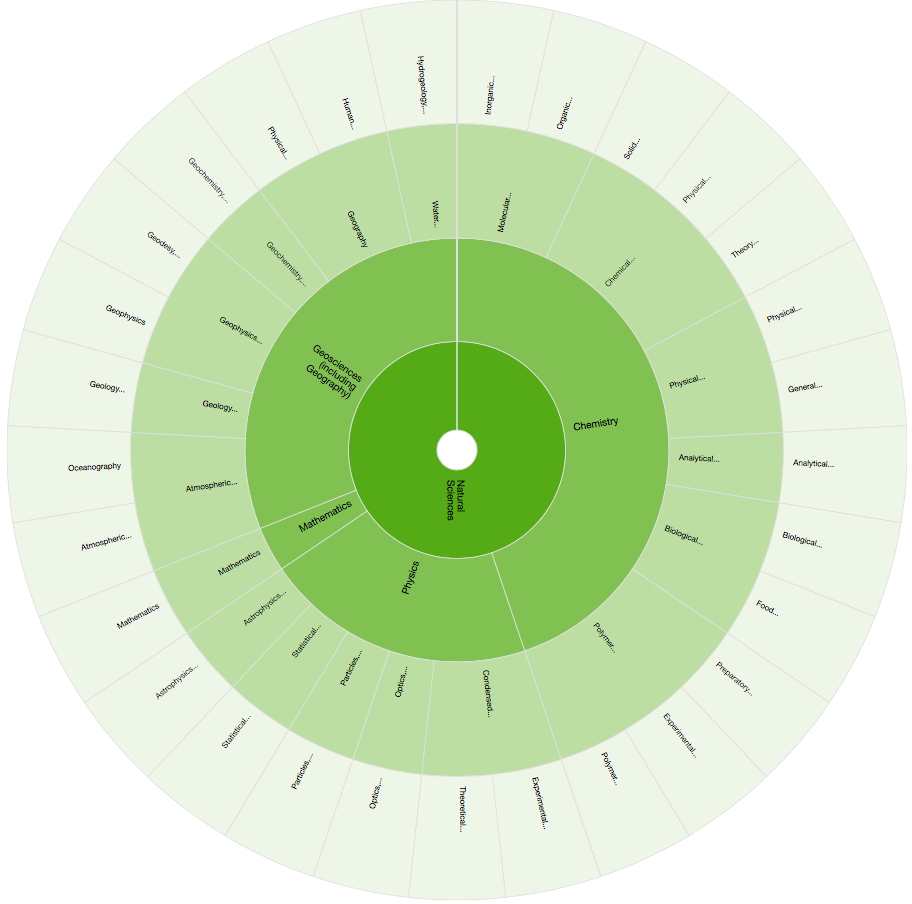
Scientists are familiar with the term data, at least in a scientific or chemical context, but appreciating metadata (meaning "after", or "beyond") is slightly more subtle, in the sense of using it to mean data about data. The challenge lies in clarifying where the boundary between data and its metadata lies and in specifying and controlling the vocabulary used for these metadata descriptions. Items in a chemical metadata dictionary might include e.g. subject classifications such as Organic Molecular Chemistry or identifiers such as InChIkey. But what could metametadata be? Here I briefly show some examples by way of illustration.
Let me start by defining a data repository as a store of both data and the metadata describing it. The metadata is to be exposed in a standard manner which allows it to be aggregated by other agencies. Nowdays, it is becoming common to identify such a data object together with its metadata using a persistent identifier, or DOI. But to decide if any particular repository and the data objects contained therein is generally useful to you, you need information about the metadata itself. Technically, this is defined using a schema[1] describing the metadata (which might e.g. identify any dictionaries used); hence metametadata. Now you need to store the metametadata and so I introduce the concept of a registry which does this. This metametadata object is itself assigned a DOI‡ and here I list these DOIs for a personal selection of some chemically oriented examples, in this case deriving from the largest registry of research data repositories re3data.org. You can search for your own entry at their site: http://service.re3data.org/search.
| Data repository | The repository metametadata DOI♣ | Badge |
|---|---|---|
| Figshare | 10.17616/R3PK5R[2] |
|
| Zenodo | 10.17616/R3QP53[3] |
|
| Cambridge structure database | 10.17616/R36011[4] |
|
| Crystallographic open database | 10.17616/R37S31[5] |
|
|
Oxford University Research Archive |
10.17616/R3Q056[6] |
|
|
Open Notebook Science |
10.17616/R3859D[7] |
|
|
Usefulchem |
10.17616/R3Z89N[8] |
|
|
Chemotion |
10.17616/R34P5T[9] |
|
|
Chemspider |
10.17616/R38P4P[10] |
|
|
Chemical Database Service |
10.17616/R36P42[11] |
|
|
Imperial College HPC data repository. |
10.17616/R3K64N[12],[13] |
|
|
Imperial College SPECTRa repository.[14] |
10.17616/R30316[15] |
|
Not all of the repositories listed in the table above assign formal DOIs to their data collections, meaning that the metadata for their entries cannot be aggregated in a searchable manner using e.g. search.datacite.org/ui (or search.datacite.org/api for the machine version). Currently, the metametadata does not fully carry this information, an aspect which I gather will be rectified in a future revision of the re3data schema.[1]
Importantly, both metadata and (repository) metametadata can be searched using APIs (application programmer interface), ensuring that the entire flow of meta information can be subject to automated software analysis rather than just visual inspections by a human.This should allow a rich and open infrastructure for handling research objects or data to be built up using hierarchical metadata. The examples above indeed show that the chemical space is already the largest component of the Natural Sciences space.
Although the edifice is still largely in its infancy, already I think we can start to see an alternative open approach emerging to "Googling" for data, or the even older traditional bespoke (i.e. non-open) services offered by commercial human-based abstractors of chemical metadata.
‡This DOI is information about the metametadata, and hence it is metametametadata, or m3data. Sorry! ♣The citations at the foot of this post are generated entirely automatically (by a WordPress plugin called Kcite) from the m3data associated with each entry, i.e. the DOI listed. Were the persistent identifier for the entry ever to be changed, this would propagate automatically to the citation, unlike the static entries in the table.
References
- J. Rücknagel, P. Vierkant, R. Ulrich, G. Kloska, E. Schnepf, D. Fichtmüller, E. Reuter, A. Semrau, M. Kindling, H. Pampel, M. Witt, F. Fritze, S. Van De Sandt, J. Klump, H. Goebelbecker, M. Skarupianski, R. Bertelmann, P. Schirmbacher, F. Scholze, C. Kramer, C. Fuchs, S. Spier, and A. Kirchhoff, "Metadata Schema for the Description of Research Data Repositories", 2015. https://doi.org/10.2312/re3.008
- Re3data.Org., "figshare", 2012. https://doi.org/10.17616/r3pk5r
- Re3data.Org., "Zenodo", 2013. https://doi.org/10.17616/r3qp53
- Re3data.Org., "The Cambridge Structural Database", 2013. https://doi.org/10.17616/r36011
- Re3data.Org., "Crystallography Open Database", 2013. https://doi.org/10.17616/r37s31
- Re3data.Org., "Oxford University Research Archive", 2014. https://doi.org/10.17616/r3q056
- Re3data.Org., "ONSchallenge", 2013. https://doi.org/10.17616/r3859d
- Re3data.Org., "UsefulChem", 2014. https://doi.org/10.17616/r3z89n
- Re3data.Org., "chemotion", 2013. https://doi.org/10.17616/r34p5t
- Re3data.Org., "ChemSpider", 2013. https://doi.org/10.17616/r38p4p
- Re3data.Org., "Chemical Database Service", 2012. https://doi.org/10.17616/r36p42
- Re3data.Org., "Imperial College Research Computing Service Data Repository", 2016. https://doi.org/10.17616/r3k64n
- H. Rzepa, "Imperial College High Performance Computing Service Data Repository Metadata Schema", 2016. https://doi.org/10.14469/hpc/382
- J. Downing, P. Murray-Rust, A.P. Tonge, P. Morgan, H.S. Rzepa, F. Cotterill, N. Day, and M.J. Harvey, "SPECTRa: The Deposition and Validation of Primary Chemistry Research Data in Digital Repositories", Journal of Chemical Information and Modeling, vol. 48, pp. 1571-1581, 2008. https://doi.org/10.1021/ci7004737
- Re3data.Org., "SPECTRa Project", 2013. https://doi.org/10.17616/r30316
Tags: Academic publishing, automated software analysis, BASE, chemical context, Chemical Database Service, chemical metadata, chemical metadata dictionary, chemical space, City: Cambridge, Data dictionary, Data management, Identifiers, Knowledge representation, programmer, Registry of Research Data Repositories, search.datacite.org/api, SPECTRa, Technology/Internet