Posts Tagged ‘Peter Murray-Rust’
Saturday, November 20th, 2010
For those of us who were around in 1985, an important chemical IT innovation occurred. We could acquire a computer which could be used to draw chemical structures in one application, and via a mysterious and mostly invisible entity called the clipboard, paste it into a word processor (it was called a Macintosh). Perchance even print the result on a laserprinter. Most students of the present age have no idea what we used to do before this innovation! Perhaps not in 1985, but at some stage shortly thereafter, and in effect without most people noticing, the return journey also started working, the so-called round trip. It seemed natural that a chemical structure diagram subjected to this treatment could still be chemically edited, and that it could make the round trip repeatedly. Little did we realise how fragile this round trip might be. Years later, the computer and its clipboard, the chemistry software, and the word processor had all moved on many generations (it is important to flag that three different vendors were involved, all using proprietary formats to weave their magic). And (on a Mac at least) the round-tripping no longer worked. Upon its return to (Chemdraw in this instance), it had been rendered inert, un-editable, and devoid of semantic meaning unless a human intervened. By the way, this process of data-loss is easily demonstrated even on this blog. The chemical diagrams you see here are similarly devoid of data, being merely bit-mapped JPG images. Which is why, on many of these posts, I put in the caption Click for 3D, which gives you access to the chemical data proper (in CML or other formats). And I throw in a digital repository identifier for good measure should you want a full dataset.
(more…)
Tags:Adobe, Apple, Apple iPad, ChemDraw 12, chemical data, chemical diagrams, chemical integrity, Chemical IT, chemical structure diagram, chemical structures, chemistry software, iPad, Mac, Mac OS X, Macintosh, Microsoft, opendata, PDF, Peter Murray-Rust, Postscript, word processor, XML
Posted in Chemical IT | 5 Comments »
Sunday, May 2nd, 2010
Peter Murray-Rust in his blog asks for examples of the Scientific Semantic Web, a topic we have both been banging on about for ten years or more (DOI: 10.1021/ci000406v). What we are seeking of course is an example of how scientific connections have been made using inference logic from semantically rich statements to be found on the Web (ideally connections that might not have previously been spotted by humans, and lie overlooked and unloved in the scientific literature). Its a tough cookie, and I look forward to the examples that Peter identifies. Meanwhile, I thought I might share here a semantically rich molecule. OK, I identified this as such not by using the Web, but as someone who is in the process of delivering an undergraduate lecture course on the topic of conformational analysis. This course takes the form of presenting a set of rules or principles which relate to the conformations of molecules, and which themselves derive from quantum mechanics, and then illustrating them with selected annotated examples. To do this, a great many semantic connections have to be made, and in the current state of play, only a human can really hope to make most of these. We really look to the semantic web as it currently is to perhaps spot a few connections that might have been overlooked in this process. So, below is a molecule, and I have made a few semantic connections for it (but have not actually fully formalised them in this blog; that is a different topic I might return to at some time). I feel in my bones that more connections could be made, and offer the molecule here as the fuse!
(more…)
Tags:chair, chemical connections, Chemical IT, chemical world, chemist, energy, Fe, General, Interesting chemistry, lowest thermodynamic free energy, organic chemist, organometallic chemist, Peter Murray-Rust, semantic web, unusual
Posted in Chemical IT, General, Interesting chemistry | 2 Comments »
Monday, September 7th, 2009
The science journal is generally acknowledged as first appearing around 1665 with the Philosophical Transactions of the Royal Society in London and (simultaneously) the French Academy of Sciences in Paris. By the turn of the millennium, around 10,000 science and medical journals were estimated to exist. By then, the Web had been around for a decade, and most journals had responded to this new medium by re-inventing themselves for it. For most part, they adopted a format which emulated paper (Acrobat), with a few embellishments (such as making the text fully searchable) and then used the Web to deliver this new reformulation of the journal. Otherwise, Robert Hooke would have easily recognized the medium he helped found in the 17th century.
(more…)
Tags:A. I. Magee, A. Jana, A. P. Dove, Acrobat, American Chemical Society, aqueous solution, Balasundaram Lavan, C. S. M Allan, C. Wentrup, Chemical IT, chemical plugin, Chemoinformatics, Colorado, D. A. Widdowson, D. C. Braddock, D. J. Williams, D. R. Carbery, D. Scheschkewitz, Dalton Trans, digital Acrobat, E. H. Smith, E. M. Barreiro, E. W. Tate, Enhance Chemical Electronic Publishing, Extrusion Reactions, F. Diederich, F. Santoro, French Academy, G. Siligardi, G. Stammler, Ge, H. S. Rzepa, HTML, I. Omlor, I. Pavlakos, Interchange Apical, Interesting chemistry, Ion-Pair Mechanisms, β-diketiminate metal alkoxides, J. Clarke, J. Jana, J. L. Arbour, J. Lorenzo Alonso-Gómez, J. P. White, J. R. Arendorf, journal editor, K. K. (Mimi) Hii, K. P. Tellmann, King, Kuok Hii, L. A. Adrio, L. Johannissen, Lewis Base Catalyst, M. E. Cass, M. Hii, M. J. Cowley, M. J. Fuchter, M. J. Harvey, M. J. Humphries, M. J. Porter, M. Jakt, M. R. Crittall, M. Ritzefeld, M. Weimar, Marshall, Michael Wright, N. Berova, N. Harada, N. J. Mason, N. Mason, N. Masumoto, O. Casher, opendata, P. G. Pringle, P. Jutzi, P. Lo, P. Seiler, Paris, Peter Murray-Rust, polymerization, Porter, printing, R. B. Moreno, R. M. Williams, R. Schleyer, R. Wilhelm, Rappaport, RDF, representative, Robert Hooke, Royal Society in London, S. Díez-González, S. Lai, S. M. Allan, S. Martin-Santamaria, Sonsoles Martên-Santamarêa, Square Pyramidal Molecules, T. Lanyon-Hogg, the Philosophical Transactions of the Royal Society, V, V(III) Co, V. C. Gibson, V. Huch, V. W. Pike, W. B. Motherwell, Web Application, Web Table, XML, XSLT, Ya-Pei Lo
Posted in Chemical IT, Interesting chemistry | 6 Comments »
Friday, April 3rd, 2009
The preceeding blog entries contain stories about chemical behaviour. If you have clicked on the diagrams, you may even have gotten a Jmol view of the relevant molecules popping up. But if you are truly curious, you may even have the urge to acquire the relevant 3D information about the molecule, and play with it yourself. Even after 15 years of the (chemical) Web, this can be distressingly difficult to achieve (or can it be that it is only myself who wishes to view molecules in their native mode?). Thus the standard mechanism is to seek out on journal pages that disarming little entry entitled supporting information and to hope that you might find something useful embedded there. Embedded is the correct description, since the information is often found within the confines of an Acrobat file, and has to be extracted from there. Indeed, that is what I had to resort to in order to write one of the blog entries below. I ground my teeth whilst doing so.
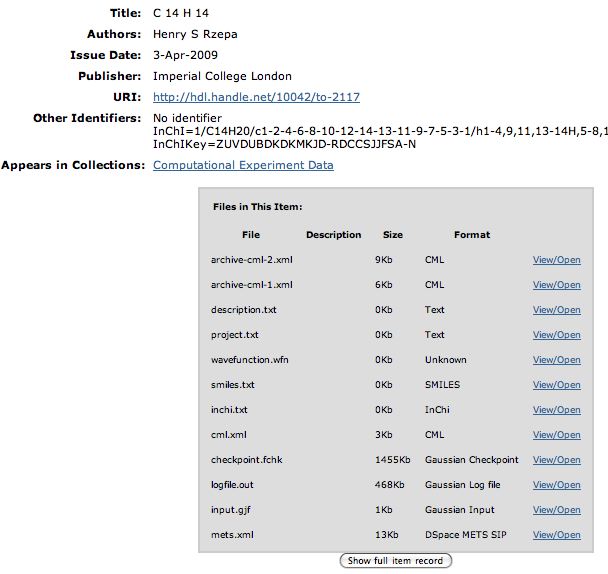
So is there a better way? We think so! The digital repository. If you click on this you should see the entry directly. What can you do there? Well, if you have suitable programs, you can download eg a Checkpoint file of the calculation that created the molecule model and re-activate it there. Or you can download just the CML file for viewing in any CML-compliant program (such as e.g. Jmol). Or you can check up on the InCHi string or the InChI Key of the molecule.
(more…)
Tags:Acrobat, Checkpoint, chemical, chemical behaviour, Chemical IT, editor in chief, opendata, Peter Murray-Rust, Web-enhanced tables
Posted in Chemical IT | No Comments »
